Frequently Asked Questions about the Rice Genome Annotation Project Resource
1. How do I cite the Rice Genome Annotation Project Resource?
2. What is source of the genomic sequence that has been annotated?
3. What is a pseudomolecule, and how were they made?
4. What is the most direct way to locate a rice sequence relative to the centromere?
5. Why are some BAC/PACs not found in pseudomolecules?
6. Do you have data files for rice intergenic sequences, 5'UTR and 3'UTR respectively?
7. How to obtain the corresponding locus ID and GenBank accession number of a gene?
9. Why are some models missing from the files on the Download site?
10. Can I order an EST clone from you?
11. Can I order a BAC/PAC clone from you?
12. How to find the coordinates for the exons and cds for each gene model?
13. How were the pseudomolecules created?
14. Which gene identifier should I use?
15. How do I download Rice Genome Annotation Project data?
16. Do you have a parser for your XML annotation format?
19. My locus identifier has disappeared in the new release. Where can I find my locus?
20. What are the steps in the automated annotation process?
21. How do I interpret the optical map data?
1. How do I refer to the Rice Genome Annotation Project (RGAP) Resource? Back to top
Researchers who wish to cite the Rice Genome Annotation Project website are encouraged to refer to these publications:
- Kawahara, Y., de la Bastide, M., Hamilton J. P., Kanamori, H., McCombie, W. R., Ouyang, S., Schwartz, D. C., Tanaka, T., Wu, J., Zhou, S., Childs, K. L., Davidson, R. M., Lin, H., Quesada-Ocampo, L., Vaillancourt, B., Sakai, H., Lee, S. S., Kim, J., Numa, H., Itoh, T., Buell, C. R., Matsumoto, T. 2013. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6:4.
- Hamilton, J. P., Li, C., Buell, C. R. 2024. The rice genome annotation project: an updated database for mining the rice genome. Nucleic Acids Research, gkae1061, https://doi.org/10.1093/nar/gkae1061
2. What is the subspecies of the rice annotated? Back to top
We use the sequence of the International Rice Genome Sequencing Project, that of Oryza sativa spp japonica cv Nipponbare.
3. What is a pseudomolecule, and how were they made? Back to top
The pseudomolecules were made by selecting an optimal tiling path of BAC and PAC clones that represent the 12 chromosomes. Overlaps between the BAC/PACs were trimmed and physical gaps are represented by a stretch of 1000 Ns. The centromeres were located due the gaps and presence of the CentO repeats. See here https://rice.uga.edu/annotation_pseudo_centromeres.shtml for the position of the centromeres. These overlapping clones were validated by comparison of the pseudomolecule sequence with the optical map for rice created by D. Schwartz of the University of Wisconsin. The pseudomolecules were error corrected using whole genome shotgun sequence of Nipponbare. Sequences from BACs and the Syngenta assembly of Nipponbare that could not be anchored are on separate pseudomolecules, the Unanchored and the Syngenta chromosomes.
Assembly of this release of the pseudomolecules, termed Os-Nipponbare-Reference-IRGSP-1.0, were made in collaboration with the Agrogenomics Research Center at the National Institute of Agrobiological Sciences, Tsukuba, Japan. This set of pseudomolecules unifies the previous MSU RGAP work with the IRGSP/RAP effort. Parallel and complementary annotation efforts by the MSU RGAP and the IRGSP/RAP continue yet are overlaid on the same set of pseudomolecules.
4. What is the most direct way to locate a rice sequence relative to the centromere? Back to top
You can use monocot centromere specific rice satellite repeat sequence CentO (GenBank accession: AF058902) to identify the centromeres on the pseudomolecules. In addition, fluorescent in situ hybridization information has also been used to map the centromeres.
5. Why are some BAC/PACs not found in pseudomolecules? Back to top
We were not able to incorporate all BAC/PAC clones into our pseudomolecules for several reasons. For example, some of the BAC/PACs are from indica, not japonica rice. Others are unfinished sequence. However, other BAC/PAC clones that are publicly available in Genbank/DDBJ/EMBL could not be incorporated into our pseudomolecules as:
- The BAC/PAC clone was released later than we finalized our current version of pseudomolecules;
- We had difficulty in mapping the BAC/PAC clone to its correct position;
- They are redundant clones and some other overlapping clones were used in the pseudomolecules in their place.
6. Do you have data files for rice intergenic sequences, 5'UTR and 3'UTR respectively? Back to top
We have files containing UTR (both 5' and 3') in our Download site when the gene model aligned well with rice EST and/or full-length cDNA sequences (see READMEREADME).
7. How to obtain the corresponding locus ID and GenBank accession number of a gene? Back to top
We have assigned pub_locus to the rice genes. They can be retrieved at the Locus Name Search. We have not submitted the annotation to GenBank unless the sequences have been finished at TIGR.
8. I noticed dates for the directories under the pseudomolecule download site have changed a few times since their release: is the database still the same? Back to top
The dates for the download site directories may not reflect the dates of data release. For each version, the README file under each directory states the date for the data release.
9. Why are some models missing from the Download site files? Back to top
We excluded the partial genes, pseudogenes, and small gene models (<50 amino acids) in the annotation files for the pseudomolecules.
10. Can I order an EST clone from you? Back to top
We do not own the EST clones. We simply obtained the sequence from public domains to build the gene index (OsGI). You can order a clone from the Japanese Ministry of Agriculture, Forestry and Fisheries (MAFF) Genome Projects or RiceGenes or The Arizona Genomics Institute.
11. Can I order a BAC/PAC clone from you? Back to top
We do not distribute the physical BAC/PAC clones. The easiest place to obtain a BAC clone would be from the The Arizona Genomics Institute or RGP. You can order individual clones online from them by using clone name as catalog number.
12. How to find the coordinates for the exons and cds for each gene model? Back to top
You can find this information in the .xml/gff3 files for genes in the pseudomolecules from the Rice Genome Annotation Project Download site.
13. How were the pseudomolecules created? Back to top
Pseudomolecules are the virtual contigs constructed by resolving discrepancies between overlapping BAC/PAC clones, trimming the overlap regions, and linking the unique sequences to form a contigous sequence for each rice chromosome. Rice pseudomolecules were generated by using a in-house, database-based, semi-automated process.
There were several key steps:
1) Overlapping regions between BACs were determined by similarity searches and tiling path information;
2) The pseudomolecules were made by removing the overlapping regions;
3) The gene models and associated features from the BAC/PAC clones were copied from the BAC/PACs to the pseudomolecules;
4) Quality control.
14. Which gene identifier should I use? Back to top
There are several gene identifiers in the Rice Genome Annotation Project database, such as locus, pub_locus, com_name, TU feat_name, model feat_name. The TU feat_name (such as 11667.t00001) and model feat_name (such as 11667.m00001) are the uid (unique identifier) for a gene in rice genome annotation database. We have implemented locus ids for Release 3. The convention for Release 3 is LOC_Osxxg##### where X is one of the 12 chromosomes and the # represents sequential loci on the pseudomolecule. Pub_locus (such as LOC_Os01g01010) is a stable identifier for genes across different versions. We encourage you to use locus id.
Please note that the feat_name for a gene in the BAC clone differs from the feat_name for the same gene in the pseudomolecule. Due to the continuous updates to most of the unfinished sequences, our automated annotations are also updated continuously and the gene identifiers will change. However, we will keep each version of our pseudomolecule releases in the database and we recommend that the users to use the dataset from pseudomolecules if you wish to trace back the gene models you are interested in. You can track your gene models among our pseudomolecule versions by using our Version Converter file.
15. How do I download Rice Genome Annotation Project annotation data? Back to top
The Rice Genome Data Download page provides a description of the formats that are available to download rice genome annotation data and a batch download tool. If you wish to download the entire data set or a number of entire chromosome data sets, please use the Download site.
16. Do you have a parser for your XML annotation format? Back to top
To retrieve each gene model and their features, you can download the following file, then unpack and unzip it: https://rice.uga.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/tools/TIGR_XML_parser.tar.gz.
Note that for Release 6.1 we have stopped using TIGR XML format and switched to distributing the annotation in validated GFF3 and tab delimited flat files.
17. What is the difference between the Rice Genome Annotation Project pseudomolecules and those of the IRGSP? Back to top
The pseudomolecules presented on our web pages and Download site have been generated as part of our National Science Foundation funded Rice Genome Annotation Project. Our pseudomolecules were constructued using all of the BAC/PAC sequences from the IRGSP which are available in Genbank/DDBJ/EMBL.
18. Is the Rice Genome Annotation Project annotation the same as that of the Rice Annotation Project (RAP)? Back to top
The Rice Genome Annotation Project and the Rice Annotation Project are separate projects focused on annotating the rice genome. Different pseudomolecules and different methods/criteria are used to identify gene models between the two groups. While some of the annotation may be similar between the two groups, the annotation will differ due to the alternative annotation methods used in the two projects. We have mapped the RAP models to our pseudomolecules to allow users to compare annotation between the two projects
19. My locus identifier has disappeared in the new release. Where can I find my locus? Back to top
Information about obsolete locus indentifiers can be found at the Obsolete Loci Search.
20. What are the steps in the automated annotation process? Back to top
The pipeline for the automated annotation process used by the Rice Genome Annotation Project can be found here.
21. How do I interpret the optical map data? Back to top
We checked the concordance of the pseudomolecules with that of the Nipponbare optical map made by the Schwartz lab https://www.ncbi.nlm.nih.gov/pubmed/17697381. The pseudomolecules were digested in silico with SwaI and aligned with the experimentally derived SwaI optical map. The alignments of all 12 pseudomolecules along with the BACs/PACs were manually examined. In general, the pseudomolecules were in good concordance with the optical map. There were 169 discordances between the optical map and the pseudomolecules. These disconcordances were classified into five classes: 1) Physical gaps in the pseudomolecules; 2) Missed or extra fragment(s). This means an extra fragment(s) either on the optical map or the pseudomolecule. Only size differences larger than 5 kb were annotated; 3) Significant size differences (mostly > 5 kb). The number of fragments are usually the same; 4) Multiple different SwaI sites in the same area; 5) Multiple different SwaI sites and multiple un-matched SwaI fragments. The total sizes are not comparable. There are possible misassemblies. The figure below shows the view of the optical map and pseudomolecules.
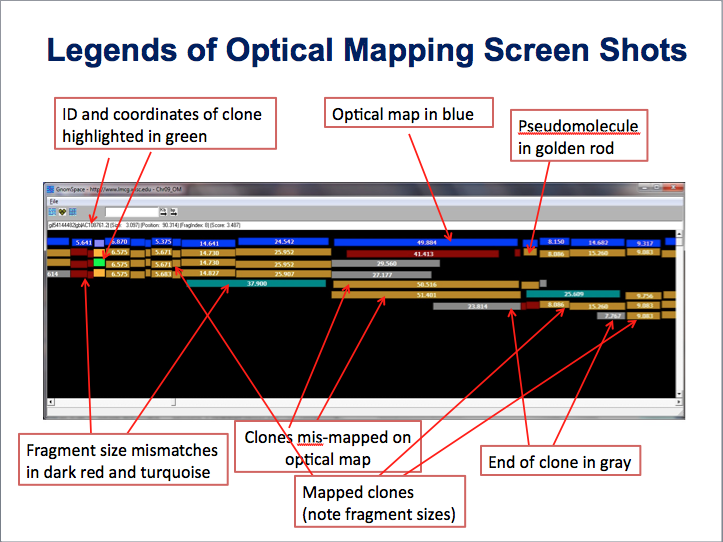

|

|

|
This work is supported by grants (DBI-0321538/DBI-0834043) from the National Science Foundation and funds from the Georgia Research Alliance, Georgia Seed Development, and University of Georgia.
